CATalog
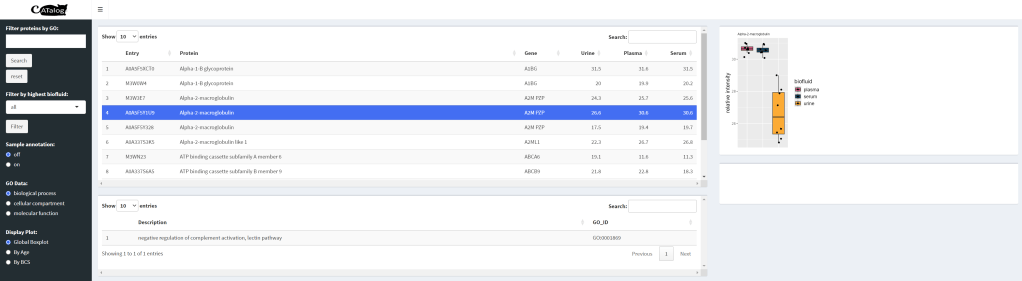
CATalog is an interactive application built in Rshiny that allows users to easily visualize protein information for the purposes of biomarker discovery. Currently, the application hosts 2000 proteins from the feline proteome. Protein data is presented in terms of relative abundance across three different biofluids Integration of gene ontology (GO) information allows for a more detailed view of each protein’s biological function, allowing for individual biomarker selection.
CATalog is currently deployed at: https://searlelab.shinyapps.io/CATalog/
InstantRamen
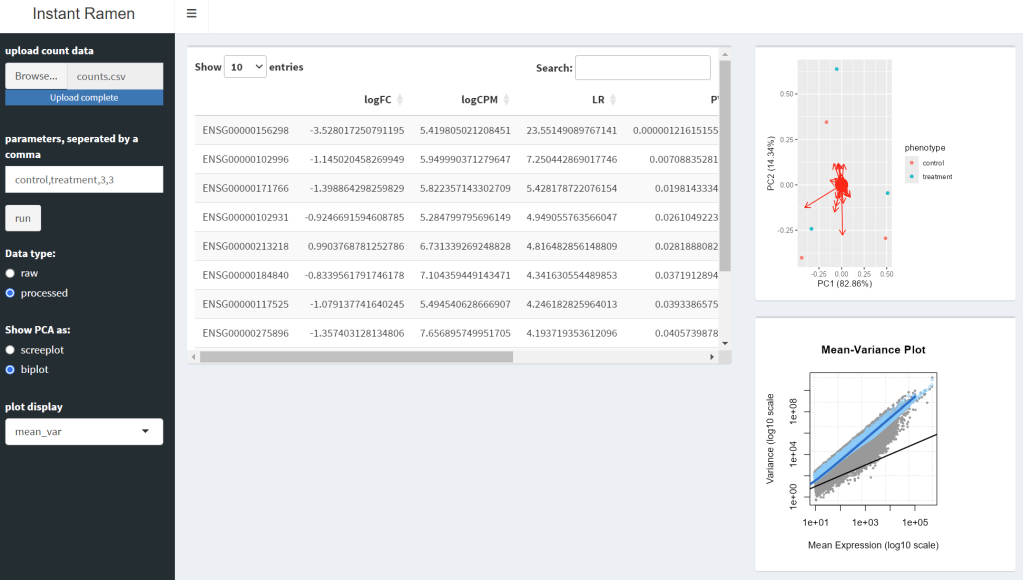
InstantRamen is an Rshiny application designed to facilitate RNAseq analysis using an EdgeR based pipeline. The program was designed with Gene Expression Omnibus (GEO) data in mind, identifying statistically significant DEGs (differently expressed genes) and visualizing the results. Additionally, annotation features are included, creating an more intuitive set of results.
InstantRamen is currently deployed at: https://alexwilliamjoyce.shinyapps.io/InstantRamen/
https://github.com/A-W-J/InstantRamen
Kinopedia
Kinopedia is a database and analysis tool for kinome array data. It was developed in the lab of Dr. Robert Smith for my master’s thesis at the University of Toledo. It was built in Rshiny and incorporates experimental, recombinant, and fingerprint data.
The current version of Kinopedia is the legacy build, which aligns with what was described in my thesis. A future, more refined version of the program is currently in development. The legacy build can be found at:
https://alexwilliamjoyce.shinyapps.io/Kinopedia–LegacyBuild/
ChemBuilder
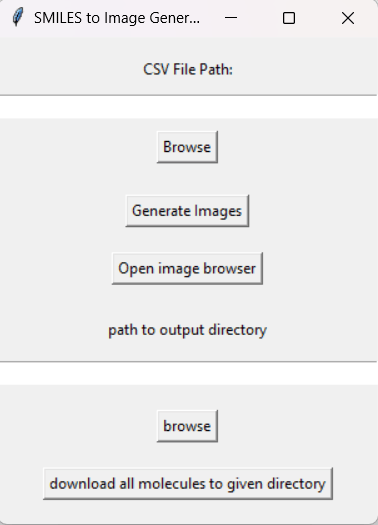
ChemBuilder is a Python-based application designed to convert SMILEs codes into images of their corresponding molecular structures from an input .csv file. The application currently uses a tkinter interface which features in-app visualization as well as a mass download feature that allows the end user to download all generated images in a single click.
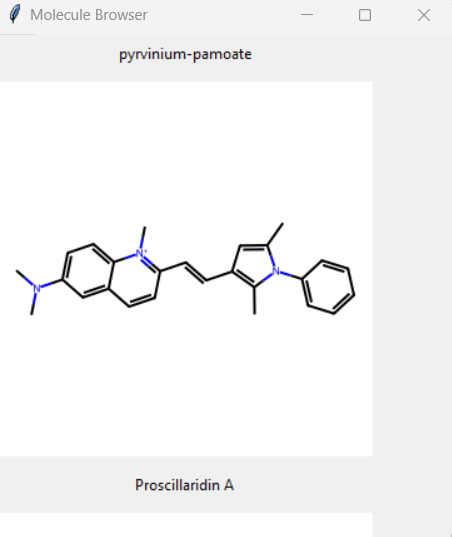
You can find ChemBuilder at: https://github.com/A-W-J/ChemBuilder
Additionally, you can download an executable version of the application from: https://drive.google.com/file/d/1LVvTjVAcYDlfHjn0iMHbnUWq3O06Oczb/view?usp=sharing
Note that the executable version of the application was created using PyIsntaller, which adds the majority of the Python backend into the .zip file. A web-based version of ChemBuilder is underway.